Plot the empirically estimated null distribution and associated p-values
Source:R/plot.R
plotNull.RdThis function takes the results from function generateNull()
and plots the density curves of permuted scores for the provided samples via
sampleNames parameter. It can plot null distribution(s) for a single
sample or multiple samples.
plotNull(
permuteResult,
scoredf,
pvals,
sampleNames = NULL,
cutoff = 0.01,
textSize = 2,
labelSize = 5
)Arguments
- permuteResult
A matrix, null distributions for each sample generated using the
generateNull()function- scoredf
A dataframe, singscores generated using the
simpleScore()function- pvals
A vector, estimated p-values using the
getPvals()functionpermuteResult,scoredfandpvalsare the results for the same samples.- sampleNames
A character vector, sample IDs for which null distributions will be plotted
- cutoff
numeric, the cutoff value for determining significance
- textSize
numeric, size of axes labels, axes values and title
- labelSize
numeric, size of label texts
Value
a ggplot object
Examples
ranked <- rankGenes(toy_expr_se)
scoredf <- simpleScore(ranked, upSet = toy_gs_up, downSet = toy_gs_dn)
# find out what backends can be registered on your machine
BiocParallel::registered()
#> $MulticoreParam
#> class: MulticoreParam
#> bpisup: FALSE; bpnworkers: 1; bptasks: 0; bpjobname: BPJOB
#> bplog: FALSE; bpthreshold: INFO; bpstopOnError: TRUE
#> bpRNGseed: 1; bptimeout: NA; bpprogressbar: FALSE
#> bpexportglobals: TRUE; bpexportvariables: FALSE; bpforceGC: FALSE
#> bpfallback: TRUE
#> bplogdir: NA
#> bpresultdir: NA
#> cluster type: FORK
#>
#> $SnowParam
#> class: SnowParam
#> bpisup: FALSE; bpnworkers: 4; bptasks: 0; bpjobname: BPJOB
#> bplog: FALSE; bpthreshold: INFO; bpstopOnError: TRUE
#> bpRNGseed: ; bptimeout: NA; bpprogressbar: FALSE
#> bpexportglobals: TRUE; bpexportvariables: TRUE; bpforceGC: FALSE
#> bpfallback: TRUE
#> bplogdir: NA
#> bpresultdir: NA
#> cluster type: SOCK
#>
#> $SerialParam
#> class: SerialParam
#> bpisup: FALSE; bpnworkers: 1; bptasks: 0; bpjobname: BPJOB
#> bplog: FALSE; bpthreshold: INFO; bpstopOnError: TRUE
#> bpRNGseed: ; bptimeout: NA; bpprogressbar: FALSE
#> bpexportglobals: FALSE; bpexportvariables: FALSE; bpforceGC: FALSE
#> bpfallback: FALSE
#> bplogdir: NA
#> bpresultdir: NA
#>
# the first one is the default backend, and it can be changed explicitly.
permuteResult = generateNull(upSet = toy_gs_up, downSet = toy_gs_dn, ranked,
B =10, seed = 1,useBPPARAM = NULL)
# call the permutation function to generate the empirical scores
#for B times.
pvals <- getPvals(permuteResult,scoredf)
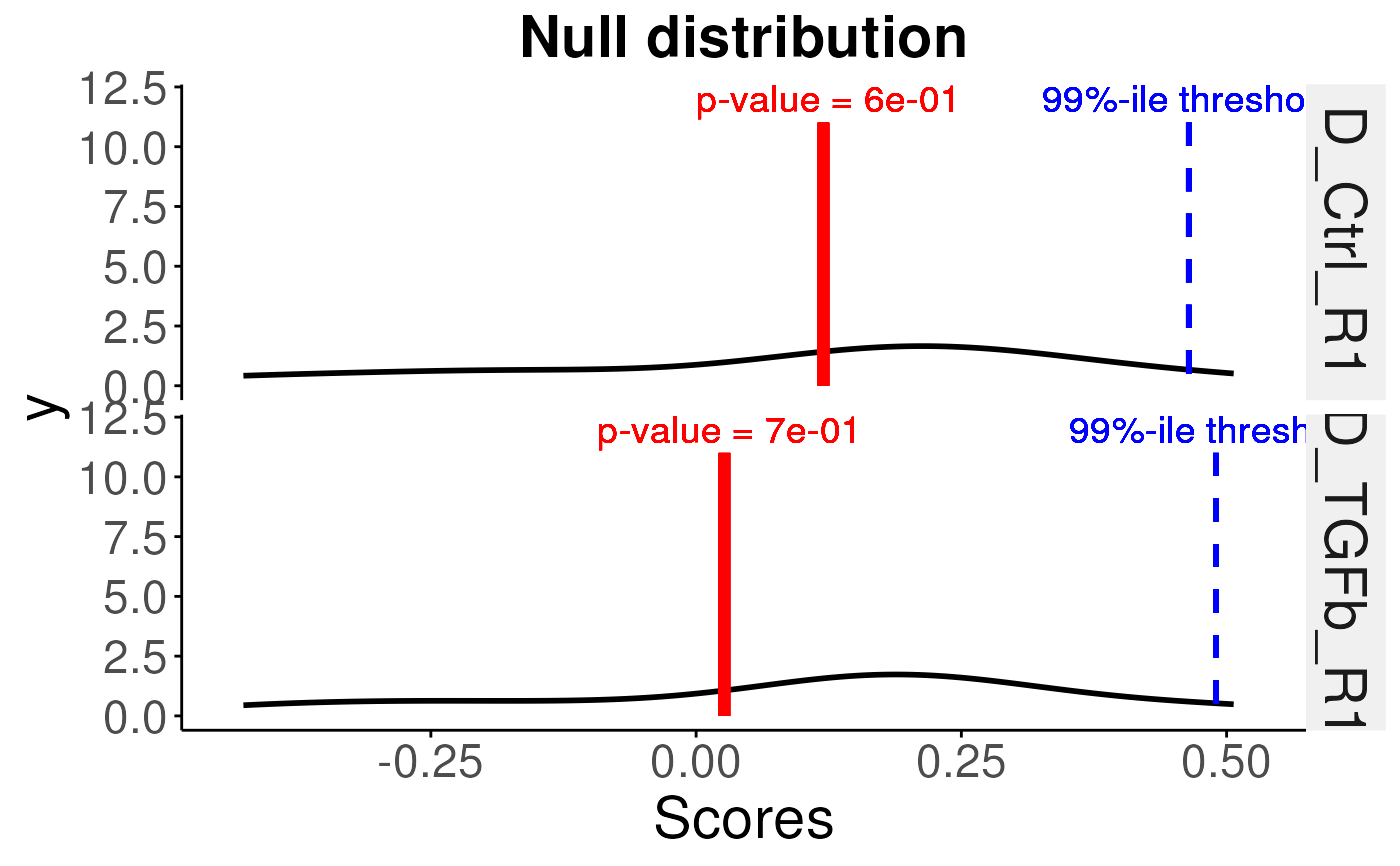
# plot for all samples
plotNull(permuteResult,scoredf,pvals,sampleNames = names(pvals))
#> Using as id variables
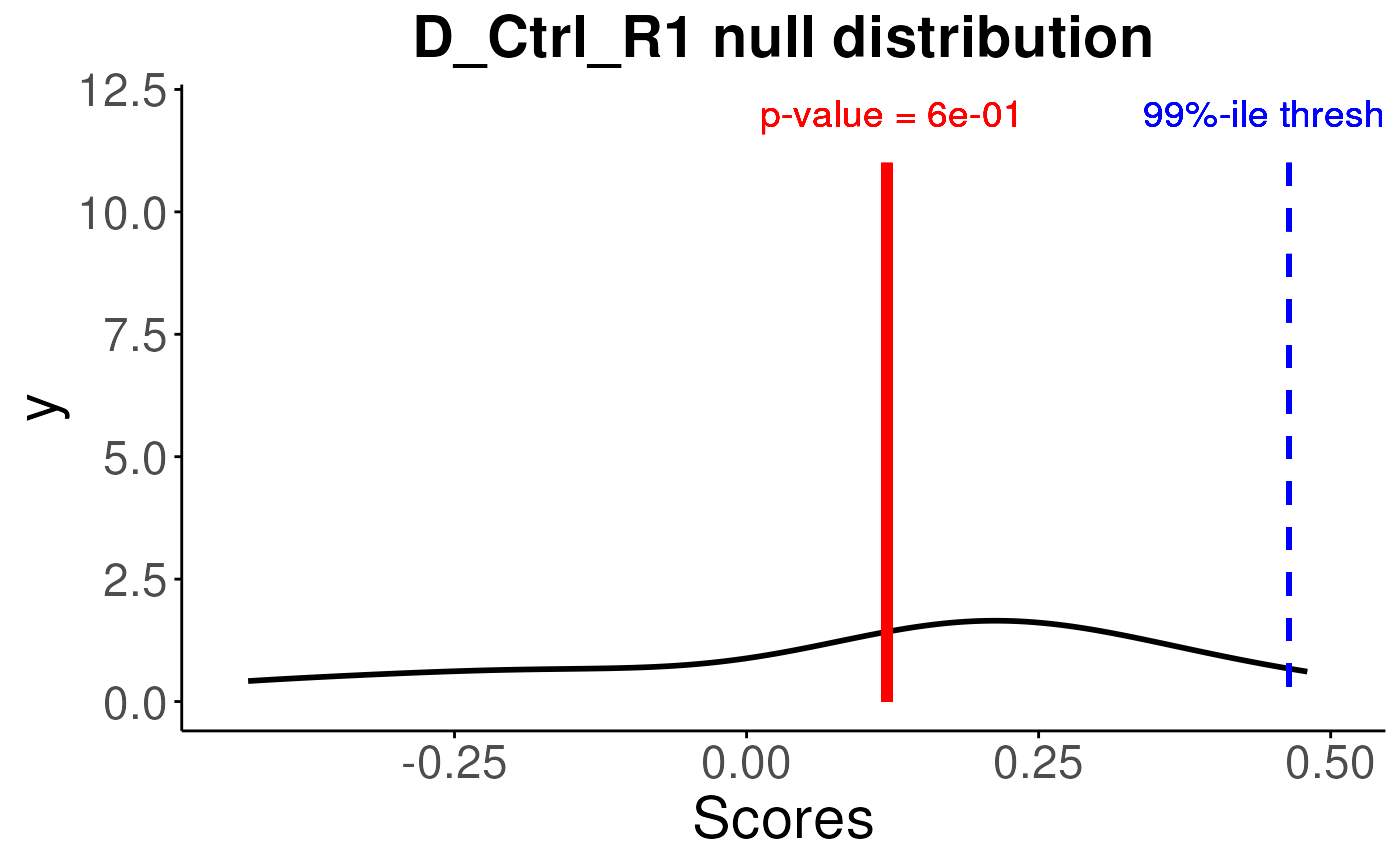
 #plot for the first sample
plotNull(permuteResult,scoredf,pvals,sampleNames = names(pvals)[1])
#plot for the first sample
plotNull(permuteResult,scoredf,pvals,sampleNames = names(pvals)[1])