plot diagnostics before and after process_data()
plot_diagnostics(expr1, expr2, group_col, abl = 2)Arguments
- expr1
expression matrix 1 for original data
- expr2
expression matrix 2 for processed data
- group_col
vector of group of samples
- abl
num, cutoff line
Value
multiple plots
Examples
data("im_data_6")
dge <- edgeR::DGEList(
counts = Biobase::exprs(im_data_6),
samples = Biobase::pData(im_data_6)
)
dge$logCPM <- edgeR::cpm(dge, log = TRUE)
proc_data <- process_data(dge,
group_col = "celltype.ch1",
target_group = "NK"
)
#> NK-Neutrophils NK-Monocytes NK-B.cells NK-CD4 NK-CD8
#> Down 4009 3944 3146 2694 2153
#> NotSig 1476 2678 4405 4985 6183
#> Up 4926 3789 2860 2732 2075
plot_diagnostics(proc_data$logCPM, proc_data$vfit$E,
group_col = proc_data$samples$group
)
#> $density
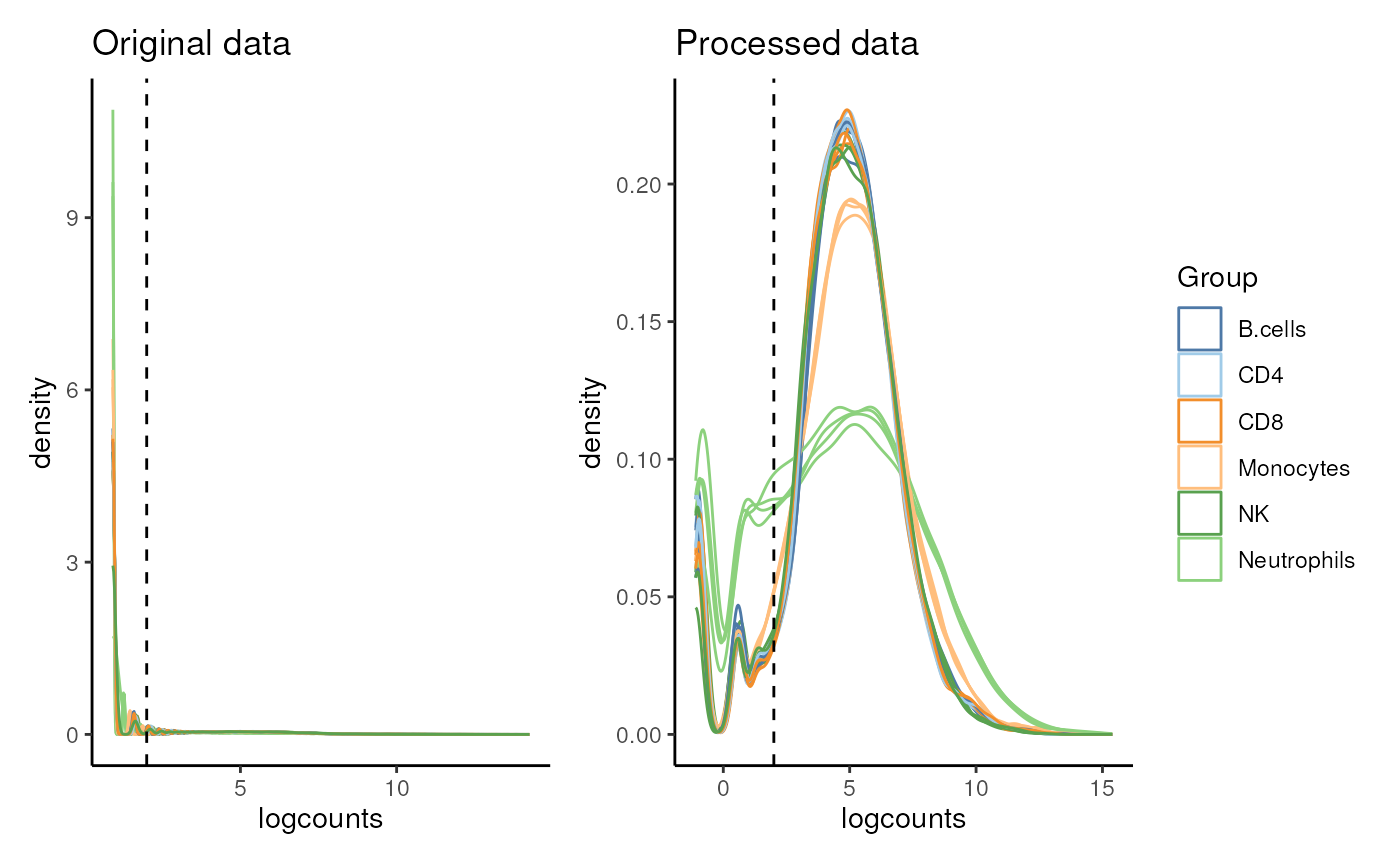 #>
#> $RLE
#>
#> $RLE
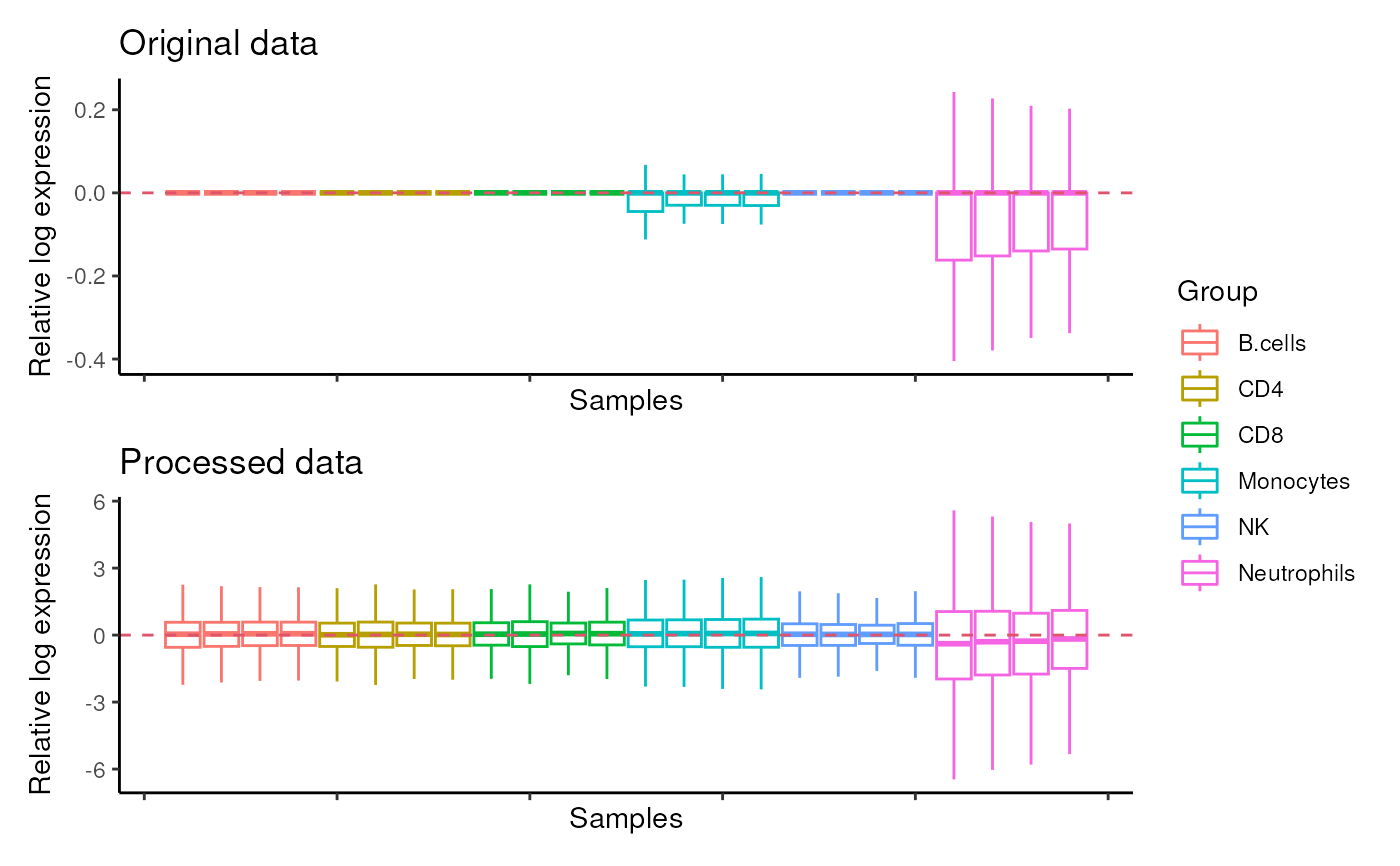 #>
#> $MDS
#>
#> $MDS
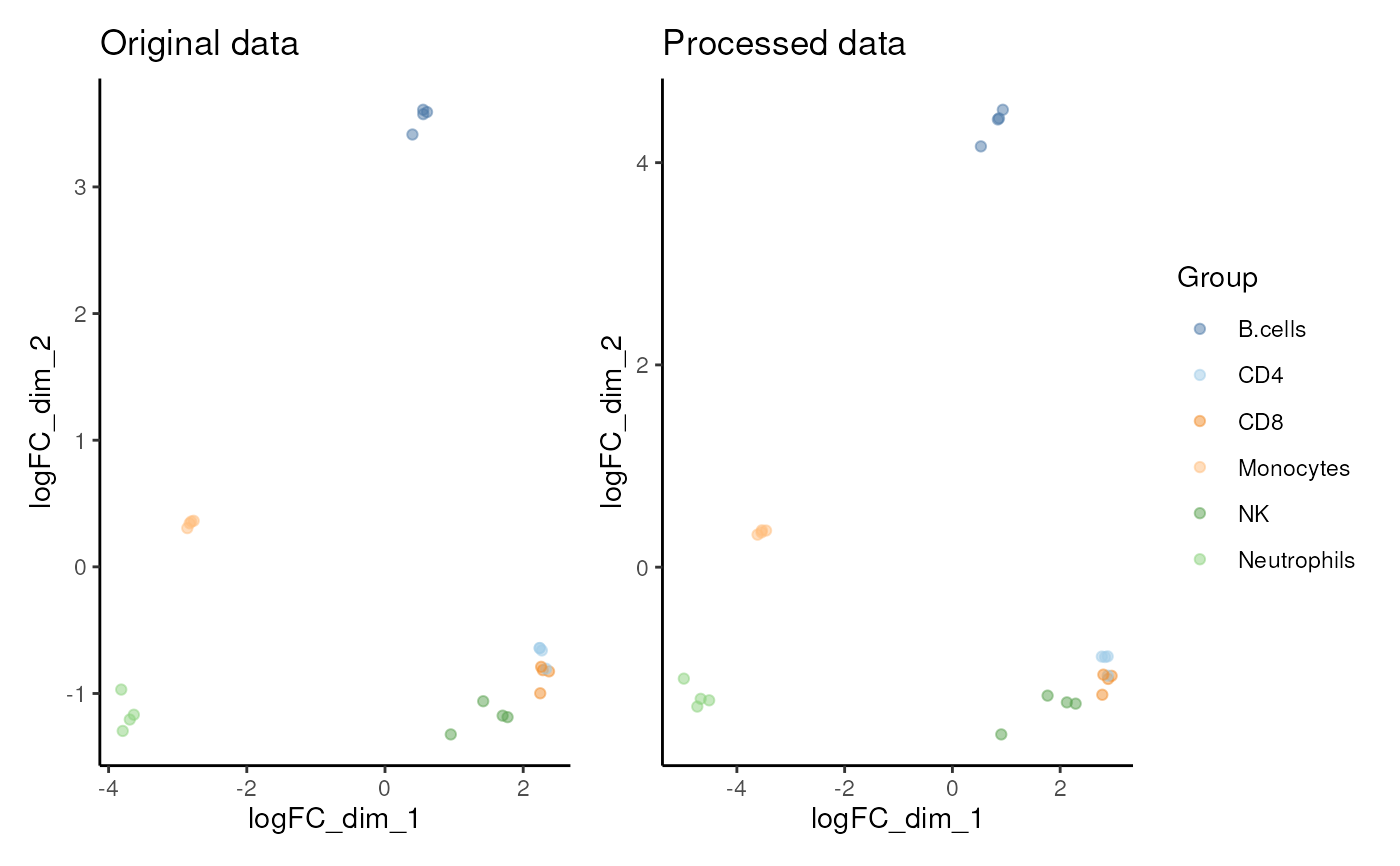 #>
#>