ExperimentList: S4 Class for handling lists of Experiments
Dharmesh D. Bhuva
15 March 2022
Source:vignettes/ExperimentList.Rmd
ExperimentList.RmdAbstract
The ExperimentList package defines S4 classes to handle data
from multiple experiments or studies by providing features of
lists as well as those of a concatenated experiment. Individual
experiments can be in the form of SummarizedExperiment, Ranged
SummarizedExperiment, SingleCellExperiment, or SpatialExperiment
objects. Annotations specific to each experiment are stored thus
providing a unified interface to dealing with data from multiple
studies. Specialised functions to access experiment data, and to
apply functions across experiments are implemented. Existing
functions implemented for each individual experiment (e.g.,
SingleCellExperiment::reducedDim()) can be readily
applied across the entire list of experiments.
Motivation
The advent of high throughput molecular measurement technologies has
resulted in the generation of vast amounts of data. The
SummarizedExperiment object and its derivates have assisted
in hosting data from these technologies. The
SingleCellExperiment and SpatialExperiment
objects are able to store even higher resolution single-cell and spatial
transcriptomics measurements from a single biological sample
respectively. Reduced costs has enabled the generation of these data
from multiple biological samples. Such data is not easily stored and
manupilated within a single object. Concatenation of objects can
partially resolve this data since it can be analysed in unison, however,
prevents for sample-wise analysis. Maintaining a list of objects would
allow object-wise analysis, but would hinder collective analysis.
The ExperimentList object is designed to fill in this
gap and allows storage and manipulation of multiple
SpatialExperiment, SingleCellExperiment,
RangedSummarizedExperiment or
SummarizedExperiment objects. It provides both list-like
and object-like functionality thus providing dynamic access to the data
as the needs arise. For example, when analysing multiple spatial
transcriptomic datasets, users may wish to compute reduced dimensions
(e.g., PCA) on each individual object before dataset integration and
compute a combined reduced dimension after. In such scenarios, having a
hybrid interface to the data is beneficial.
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("ExperimentList")Anatomy of an ExperimentList
The ExperimentList object borrows its structure from the
object it hosts. When dealing with a list of
SpatialExperiment objects, all slots from the object are
made available therefore the rows contain feature (e.g.,
gene/transcript) information, the columns represent individual
observations (e.g., visium spots), and multiple assays can be stored.
The individual objects (different shades in the schematic) are
concatenated to create a single SpatialExperiment object.
The mapping between the original sample and the concatenated columns are
maintained using and internal slot (experimentIndex).
Sample-specific annotation (NOT spot-specific) are held in a
DataFrame and are linked to the columns of the new
ExperimentList object.
The following schematic demonstrates the structure of the ExperimentList object and lists some common accessors to interact with the object.
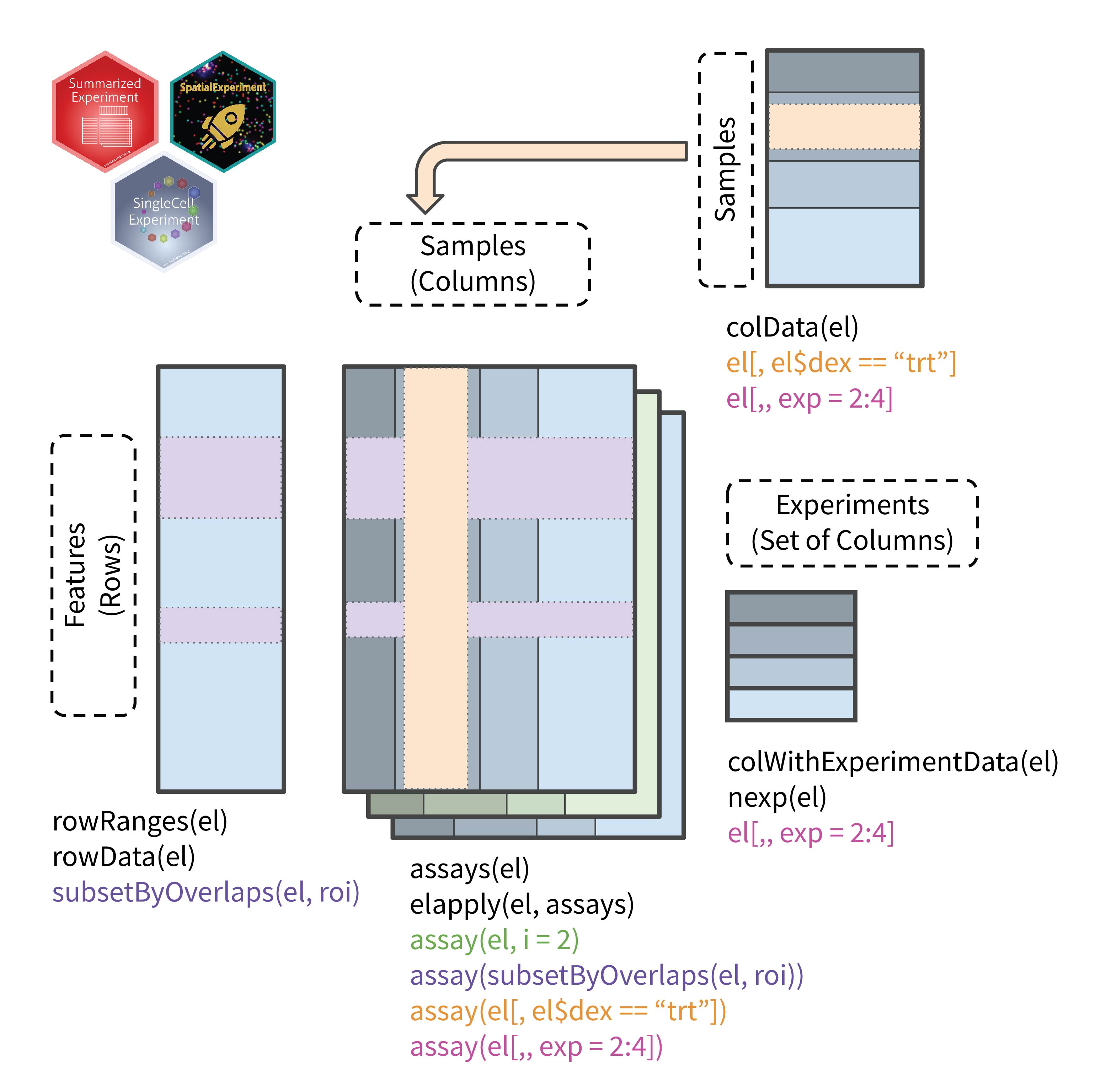
ExperimentList anatomy
Under the hood, a separate class is used to hold the list of objects as described in the table below.
| Object class | Matched ExperimentList class |
|---|---|
SummarizedExperiment |
SummarizedExperimentList |
RangedSummarizedExperiment |
RangedSummarizedExperimentList |
SingleCellExperiment |
SingleCellExperimentList |
SpatialExperiment |
SpatialExperimentList |
Constructing an ExperimentList
The TENxVisiumData package contains 10X Visium data from
various human and mouse tissues. We will use the breast cancer IDC and
ILC datasets from this package to demonstrate the ExperimentList
container. These data contain measurements of 36601 transcripts measured
across 7785 and 4325 spots respectively.
library(ExperimentList)
library(TENxVisiumData)
#download data
spe1 = TENxVisiumData::HumanBreastCancerIDC()
spe2 = TENxVisiumData::HumanBreastCancerILC()
#remove alt exps - these should be matched across exps (likewise for rownames)
altExps(spe2) = list()
#create a list of objects
spe_list = list(
'HumanBreastCancerIDC' = spe1,
'HumanBreastCancerILC' = spe2
)
spe_list
#> $HumanBreastCancerIDC
#> class: SpatialExperiment
#> dim: 36601 7785
#> metadata(0):
#> assays(1): counts
#> rownames(36601): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
#> ENSG00000277196
#> rowData names(1): symbol
#> colnames(7785): AAACAAGTATCTCCCA-1 AAACACCAATAACTGC-1 ...
#> TTGTTTGTATTACACG-1 TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#>
#> $HumanBreastCancerILC
#> class: SpatialExperiment
#> dim: 36601 4325
#> metadata(0):
#> assays(1): counts
#> rownames(36601): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
#> ENSG00000277196
#> rowData names(1): symbol
#> colnames(4325): AAACAACGAATAGTTC-1 AAACAAGTATCTCCCA-1 ...
#> TTGTTTCCATACAACT-1 TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactorGiven the individual objects, we first create a sample-specific
annotation table and subsequently use it to create an
ExperimentList object. With the data prepared, the object
can be constructed using the ExperimentList() function.
#create some artificial experiment annotations
experimentAnnotation = data.frame(
sex = c('Female', 'Female'),
age = c(65, 68),
row.names = c('HumanBreastCancerIDC', 'HumanBreastCancerILC')
)
experimentAnnotation
#> sex age
#> HumanBreastCancerIDC Female 65
#> HumanBreastCancerILC Female 68
#create ExperimentList objects
el = ExperimentList(experiments = spe_list, experimentData = experimentAnnotation)
el
#> ExperimentList with 2 SpatialExperiments
#> class: SpatialExperimentList
#> dim: 36601 12110
#> metadata(0):
#> assays(1): counts
#> rownames(36601): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
#> ENSG00000277196
#> rowData names(1): symbol
#> colnames(12110): HumanBreastCancerIDC.AAACAAGTATCTCCCA-1
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 ...
#> HumanBreastCancerILC.TTGTTTCCATACAACT-1
#> HumanBreastCancerILC.TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#> experiments: 2
#> experimentNames (2): HumanBreastCancerIDC HumanBreastCancerILC
#> experimentData names (2): sex ageThe ExperimentList object can be created without
experimentData and with an unnamed list. When using a named
list, names of the list should be matched with rownames of
experimentData.
Common operations on ExperimentList
Subsetting
Subsetting can be performed in a similar way to the parental
SummarizedExperiment object using the [
function.
#subset the first five features and first three samples
el[1:5, 1:3]
#> ExperimentList with 2 SpatialExperiments
#> class: SpatialExperimentList
#> dim: 5 3
#> metadata(0):
#> assays(1): counts
#> rownames(5): ENSG00000243485 ENSG00000237613 ENSG00000186092
#> ENSG00000238009 ENSG00000239945
#> rowData names(1): symbol
#> colnames(3): HumanBreastCancerIDC.AAACAAGTATCTCCCA-1
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1
#> HumanBreastCancerIDC.AAACAGAGCGACTCCT-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#> experiments: 2
#> experimentNames (2): HumanBreastCancerIDC HumanBreastCancerILC
#> experimentData names (2): sex ageIn addition to this, entire experiments can be subsetted using the
[ function as below.
#subset the first five features and all columns from the second experiment
el[1:5, , exp = 2]
#> ExperimentList with 1 SpatialExperiments
#> class: SpatialExperimentList
#> dim: 5 4325
#> metadata(0):
#> assays(1): counts
#> rownames(5): ENSG00000243485 ENSG00000237613 ENSG00000186092
#> ENSG00000238009 ENSG00000239945
#> rowData names(1): symbol
#> colnames(4325): HumanBreastCancerILC.AAACAACGAATAGTTC-1
#> HumanBreastCancerILC.AAACAAGTATCTCCCA-1 ...
#> HumanBreastCancerILC.TTGTTTCCATACAACT-1
#> HumanBreastCancerILC.TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#> experiments: 1
#> experimentNames (1): HumanBreastCancerILC
#> experimentData names (2): sex ageGetters and setters
The functions below can be used to access and set data in the object.
#number of experiments
nexp(el)
#> [1] 2
#names of experiments
experimentNames(el)
#> [1] "HumanBreastCancerIDC" "HumanBreastCancerILC"
#get a list of individual experiments
experiments(el)
#> $HumanBreastCancerIDC
#> class: SpatialExperiment
#> dim: 36601 7785
#> metadata(0):
#> assays(1): counts
#> rownames(36601): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
#> ENSG00000277196
#> rowData names(1): symbol
#> colnames(7785): AAACAAGTATCTCCCA-1 AAACACCAATAACTGC-1 ...
#> TTGTTTGTATTACACG-1 TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#>
#> $HumanBreastCancerILC
#> class: SpatialExperiment
#> dim: 36601 4325
#> metadata(0):
#> assays(1): counts
#> rownames(36601): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
#> ENSG00000277196
#> rowData names(1): symbol
#> colnames(4325): AAACAACGAATAGTTC-1 AAACAAGTATCTCCCA-1 ...
#> TTGTTTCCATACAACT-1 TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#get experiment annotations
experimentData(el)
#> DataFrame with 2 rows and 2 columns
#> sex age
#> <character> <numeric>
#> HumanBreastCancerIDC Female 65
#> HumanBreastCancerILC Female 68
#get column annotations
head(colData(el))
#> DataFrame with 6 rows and 1 column
#> sample_id
#> <character>
#> HumanBreastCancerIDC.AAACAAGTATCTCCCA-1 HumanBreastCancerIDC1
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 HumanBreastCancerIDC1
#> HumanBreastCancerIDC.AAACAGAGCGACTCCT-1 HumanBreastCancerIDC1
#> HumanBreastCancerIDC.AAACAGGGTCTATATT-1 HumanBreastCancerIDC1
#> HumanBreastCancerIDC.AAACAGTGTTCCTGGG-1 HumanBreastCancerIDC1
#> HumanBreastCancerIDC.AAACATTTCCCGGATT-1 HumanBreastCancerIDC1
#get column annotations merged with experiment annotations
head(colData(el, experimentData = TRUE))
#> DataFrame with 6 rows and 3 columns
#> sample_id sex
#> <character> <character>
#> HumanBreastCancerIDC.AAACAAGTATCTCCCA-1 HumanBreastCancerIDC1 Female
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 HumanBreastCancerIDC1 Female
#> HumanBreastCancerIDC.AAACAGAGCGACTCCT-1 HumanBreastCancerIDC1 Female
#> HumanBreastCancerIDC.AAACAGGGTCTATATT-1 HumanBreastCancerIDC1 Female
#> HumanBreastCancerIDC.AAACAGTGTTCCTGGG-1 HumanBreastCancerIDC1 Female
#> HumanBreastCancerIDC.AAACATTTCCCGGATT-1 HumanBreastCancerIDC1 Female
#> age
#> <numeric>
#> HumanBreastCancerIDC.AAACAAGTATCTCCCA-1 65
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 65
#> HumanBreastCancerIDC.AAACAGAGCGACTCCT-1 65
#> HumanBreastCancerIDC.AAACAGGGTCTATATT-1 65
#> HumanBreastCancerIDC.AAACAGTGTTCCTGGG-1 65
#> HumanBreastCancerIDC.AAACATTTCCCGGATT-1 65Apply
The elapply() function can be used to apply functions to
individual objects.
#apply function
elapply(el, dim)
#> $HumanBreastCancerIDC
#> [1] 36601 7785
#>
#> $HumanBreastCancerILC
#> [1] 36601 4325If the return type is the same as the type of the individual
experiment objects, they will be combined into
ExperimentList object.
#get the first 100 spots
elapply(el, function(x) x[, 1:100])
#> ExperimentList with 2 SpatialExperiments
#> class: SpatialExperimentList
#> dim: 36601 200
#> metadata(0):
#> assays(1): counts
#> rownames(36601): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
#> ENSG00000277196
#> rowData names(1): symbol
#> colnames(200): HumanBreastCancerIDC.AAACAAGTATCTCCCA-1
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 ...
#> HumanBreastCancerILC.AACCCGAGCAGAATCG-1
#> HumanBreastCancerILC.AACCCTACTGTCAATA-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#> experiments: 2
#> experimentNames (2): HumanBreastCancerIDC HumanBreastCancerILC
#> experimentData names (2): sex ageThis function allows the ExperimentList object to be
treated as a list. Since the parent class of the object is
SpatialExperiment, all functions that work for it
automatically work for the ExperimentList object.
#extract image data for each object separately
elapply(el, imgData)
#> $HumanBreastCancerIDC
#> DataFrame with 2 rows and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 HumanBreastCancerIDC1 lowres #### 0.0247525
#> 2 HumanBreastCancerIDC2 lowres #### 0.0247525
#>
#> $HumanBreastCancerILC
#> DataFrame with 1 row and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 HumanBreastCancerILC.. lowres #### 0.0247525
#extract image data collectively
imgData(el)
#> DataFrame with 3 rows and 4 columns
#> sample_id image_id data scaleFactor
#> <character> <character> <list> <numeric>
#> 1 HumanBreastCancerIDC1 lowres #### 0.0247525
#> 2 HumanBreastCancerIDC2 lowres #### 0.0247525
#> 3 HumanBreastCancerILC.. lowres #### 0.0247525Coercion
ExperimentList objects can be coerced to their parental
classes or ExperimentList versions of their parental
classes.
#convert to SpatialExperiment
as(el, 'SpatialExperiment')
#> class: SpatialExperiment
#> dim: 36601 12110
#> metadata(0):
#> assays(1): counts
#> rownames(36601): ENSG00000243485 ENSG00000237613 ... ENSG00000278817
#> ENSG00000277196
#> rowData names(1): symbol
#> colnames(12110): HumanBreastCancerIDC.AAACAAGTATCTCCCA-1
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 ...
#> HumanBreastCancerILC.TTGTTTCCATACAACT-1
#> HumanBreastCancerILC.TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> reducedDimNames(0):
#> mainExpName: NULL
#> altExpNames(0):
#> spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
#> imgData names(4): sample_id image_id data scaleFactor
#convert to SummarizedExperiment
as(el, 'SummarizedExperiment')
#> class: SummarizedExperiment
#> dim: 36601 12110
#> metadata(0):
#> assays(1): counts
#> rownames: NULL
#> rowData names(0):
#> colnames(12110): HumanBreastCancerIDC.AAACAAGTATCTCCCA-1
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 ...
#> HumanBreastCancerILC.TTGTTTCCATACAACT-1
#> HumanBreastCancerILC.TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#convert to SummarizedExperimentList
as(el, 'SummarizedExperimentList')
#> ExperimentList with 2 SummarizedExperiments
#> class: SummarizedExperimentList
#> dim: 36601 12110
#> metadata(0):
#> assays(1): counts
#> rownames: NULL
#> rowData names(0):
#> colnames(12110): HumanBreastCancerIDC.AAACAAGTATCTCCCA-1
#> HumanBreastCancerIDC.AAACACCAATAACTGC-1 ...
#> HumanBreastCancerILC.TTGTTTCCATACAACT-1
#> HumanBreastCancerILC.TTGTTTGTGTAAATTC-1
#> colData names(1): sample_id
#> experiments: 2
#> experimentNames (2): HumanBreastCancerIDC HumanBreastCancerILC
#> experimentData names (2): sex ageThe full coercion hierarchy can be explored using the
is() function.
is(el)
#> [1] "SpatialExperimentList" "SpatialExperiment"
#> [3] "ExperimentList" "SingleCellExperiment"
#> [5] "RangedSummarizedExperiment" "SummarizedExperiment"
#> [7] "RectangularData" "Vector"
#> [9] "Annotated" "vector_OR_Vector"Session information
sessionInfo()
#> R Under development (unstable) (2022-03-10 r81874)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 20.04.4 LTS
#>
#> Matrix products: default
#> BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
#>
#> locale:
#> [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
#> [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
#> [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] TENxVisiumData_1.3.0 ExperimentHub_2.3.5
#> [3] AnnotationHub_3.3.9 BiocFileCache_2.3.4
#> [5] dbplyr_2.1.1 ExperimentList_0.99.0
#> [7] SpatialExperiment_1.5.4 SingleCellExperiment_1.17.2
#> [9] SummarizedExperiment_1.25.3 Biobase_2.55.0
#> [11] GenomicRanges_1.47.6 GenomeInfoDb_1.31.4
#> [13] IRanges_2.29.1 MatrixGenerics_1.7.0
#> [15] matrixStats_0.61.0 S4Vectors_0.33.10
#> [17] BiocGenerics_0.41.2
#>
#> loaded via a namespace (and not attached):
#> [1] rjson_0.2.21 ellipsis_0.3.2
#> [3] rprojroot_2.0.2 scuttle_1.5.0
#> [5] XVector_0.35.0 fs_1.5.2
#> [7] bit64_4.0.5 interactiveDisplayBase_1.33.0
#> [9] AnnotationDbi_1.57.1 fansi_1.0.2
#> [11] R.methodsS3_1.8.1 sparseMatrixStats_1.7.0
#> [13] cachem_1.0.6 knitr_1.37
#> [15] jsonlite_1.8.0 png_0.1-7
#> [17] R.oo_1.24.0 shiny_1.7.1
#> [19] HDF5Array_1.23.2 BiocManager_1.30.16
#> [21] compiler_4.2.0 httr_1.4.2
#> [23] dqrng_0.3.0 assertthat_0.2.1
#> [25] Matrix_1.4-0 fastmap_1.1.0
#> [27] limma_3.51.5 cli_3.2.0
#> [29] later_1.3.0 htmltools_0.5.2
#> [31] tools_4.2.0 glue_1.6.2
#> [33] GenomeInfoDbData_1.2.7 dplyr_1.0.8
#> [35] rappdirs_0.3.3 Rcpp_1.0.8.2
#> [37] jquerylib_0.1.4 pkgdown_2.0.2.9000
#> [39] vctrs_0.3.8 Biostrings_2.63.1
#> [41] rhdf5filters_1.7.0 DelayedMatrixStats_1.17.0
#> [43] xfun_0.30 stringr_1.4.0
#> [45] beachmat_2.11.0 mime_0.12
#> [47] lifecycle_1.0.1 edgeR_3.37.0
#> [49] zlibbioc_1.41.0 BiocStyle_2.23.1
#> [51] ragg_1.2.2 promises_1.2.0.1
#> [53] parallel_4.2.0 rhdf5_2.39.6
#> [55] yaml_2.3.5 curl_4.3.2
#> [57] memoise_2.0.1 sass_0.4.0
#> [59] stringi_1.7.6 RSQLite_2.2.10
#> [61] BiocVersion_3.15.0 desc_1.4.1
#> [63] filelock_1.0.2 BiocParallel_1.29.17
#> [65] rlang_1.0.2 pkgconfig_2.0.3
#> [67] systemfonts_1.0.4 bitops_1.0-7
#> [69] evaluate_0.15 lattice_0.20-45
#> [71] purrr_0.3.4 Rhdf5lib_1.17.3
#> [73] bit_4.0.4 tidyselect_1.1.2
#> [75] magrittr_2.0.2 R6_2.5.1
#> [77] magick_2.7.3 generics_0.1.2
#> [79] DelayedArray_0.21.2 DBI_1.1.2
#> [81] withr_2.5.0 pillar_1.7.0
#> [83] prettydoc_0.4.1 KEGGREST_1.35.0
#> [85] RCurl_1.98-1.6 tibble_3.1.6
#> [87] crayon_1.5.0 DropletUtils_1.15.2
#> [89] utf8_1.2.2 rmarkdown_2.13
#> [91] locfit_1.5-9.5 grid_4.2.0
#> [93] blob_1.2.2 digest_0.6.29
#> [95] xtable_1.8-4 httpuv_1.6.5
#> [97] R.utils_2.11.0 textshaping_0.3.6
#> [99] bslib_0.3.1